INTRODUCTION
Interleukin-6 (IL-6), a pleiotropic cytokine that mediates many physiological and pathophysiological processes, is often regarded as a pro-inflammatory cytokine secreted by inflammatory cells, with elevated plasma concentrations observed in conditions where inflammation is present [1]. Conversely, although exercise-associated IL-6 secretion from leukocytes is negligible [2], IL-6 is released as a “myokine” from contracting muscles to enable muscle-organ crosstalk, which is important in the body’s systemic anti-inflammatory response to exercise [1,2].
IL-6’s apparently opposing actions may be attributed to its divergent signalling pathways. IL-6 directly signals through its cell-surface receptor (membrane-bound IL-6R [‘mbIL-6R’]), which initiates downstream intracellular responses via glycoprotein-130 (‘gp130’) [3,4]. This pathway, labelled “classical signalling”, only occurs in mbIL-6R-expressing cell-types (eg. myocytes, neutrophils, monocyte-macrophages) [3,4]. However, mbIL-6R can be cleaved into two fragments, one of which is lost from cells as soluble IL-6R [‘sIL-6R’]) [3,4]. Circulating IL-6 binds to sIL-6R; the resulting IL-6/sIL-6R “active complex” can bind to gp130 (which is ubiquitously expressed, but which does not bind to IL-6 alone, or sIL-6R alone) [3], and trigger “trans-signalling” responses independent of mbIL-6R [4]. Finally, a circulating soluble fragment of gp130 (‘sgp130’) binds to IL-6/sIL-6R, forming an “inactive (or buffer) complex” and impairing trans-signalling [3]. Thus, whether IL-6 signals via classical or trans-signalling is determined by the respective levels of IL-6, sIL-6R, and sgp130.
The IL-6R gene contains a single clinically-relevant single-nucleotide-polymorphism (‘SNP’): A→C at nucleotide position-1073 (NCBI Accession Code rs2228145) [5]. This results in an Aspartate→Alanine change at position-358 in the IL-6R protein (adjacent to the IL-6R cleavage site); approximately two-fold increases in cleavage result, with rs2228145 being responsible for ~50% of the variability in blood-borne sIL-6R levels [5].
Neutralisation of IL-6 trans-signalling using anti-sIL-6R antibody therapy has been approved as an effective treatment of rheumatoid arthritis [6] (and is currently undergoing clinical trials regarding the treatment of asthma [7] and COVID-19 [8]). rs2228145 is linked with several inflammatory conditions [9,10], and it seems plausible that rs2228145-inflammatory disease associations may be linked to excessive pro-inflammatory trans-signalling due to elevated sIL-6R levels, particularly in individuals possessing the CC genotype. We have previously reported that blood-borne sIL-6R levels are positively associated with adverse reactions (upper respiratory symptoms, perceived stress, worse mood, fatigue, poor sleep-quality – collectively referred to as ‘over-reaching’) in athletes undertaking prolonged periods of intense/high-volume exercise training [11]. Therefore, we hypothesised that rs2228145 may, via its impact on blood-borne sIL-6R levels, influence one’s ability to tolerate physical activity (PA) without experiencing adverse effects. Accordingly, we aimed to undertake preliminary studies exploring the relationships between rs2228145 genotype; blood-borne sIL-6R, IL-6/sIL-6R and sgp130; and self-selected PA levels in healthy adults.
METHODS
Ethics/Governance/Participant Recruitment
In this exploratory pilot study, two small participant cohorts were recruited from staff/students at Cardiff Metropolitan University. Exclusion criteria were underlying inflammatory disease, or circumstances preventing normal habitual PA levels.
Group-1: 12 participants (8 male; 4 female; age=26±9yrs) donated one set of samples each.
Group-2: 6 participants (4 male; 2 female; age=39±14yrs) underwent a longitudinal study, donating samples via five sampling points over a period of several months.
Ethical approval was obtained from Cardiff Metropolitan University School of Sport and Health Sciences ethics committee; all procedures conformed to the Declaration of Helsinki.
Sample Collection/Processing.
All participants refrained from undertaking exercise for >12h before sample donation. Capillary blood was collected from the fingertip using heparinized microvette capillary blood collection-tubes (Sarstedt, Numbrecht, Germany). The resulting samples were fractionated using a double-centrifugation method (2x1min; 6000rpm) to obtain acellular plasma samples, which were stored at -80ºC for subsequent batched analysis. To collect DNA samples, each participant swilled 10ml ddH2O around his/her mouth for 1min, before spitting the contents into a tube. After centrifugation (3000rpm; 3min), pellets (containing buccal epithelial cells) were resuspended in 350µl Chelex/4µl Proteinase K (10ng/µl; both from Sigma, Poole, UK). Samples were incubated (56oC; 30min), heated (98oC; 15min), and centrifuged (13000rpm; 3min). The buccal epithelial cell DNA content in the resulting supernatant samples was determined using Nanodrop spectrophotometry (Fisher-Scientific, Loughborough, UK); samples were stored at -80oC for subsequent batched analysis.
ELISA
Duoset ELISA kits (dy227, dy8139, dy228; R&D Systems, Abingdon, UK) were used according to manufacturers’ instructions to determine sIL-6R, IL-6/sIL-6R complex, and sgp130 levels respectively, in plasma samples (diluted 1:100 v/v, to bring readings within the range of the standard curve provided). Inter-assay coefficients of variability were calculated from readings obtained from different ELISA plates for each experiment, and were found to be <15%. Post-hoc analyses determined ratios of [IL-6/sIL-6R]:[total sIL-6R] and [total sIL-6R]:[sgp130] in each case, to estimate proportions of sIL-6R in ‘active complexes’ or ‘inactive complexes’, respectively.
Genotyping Assay
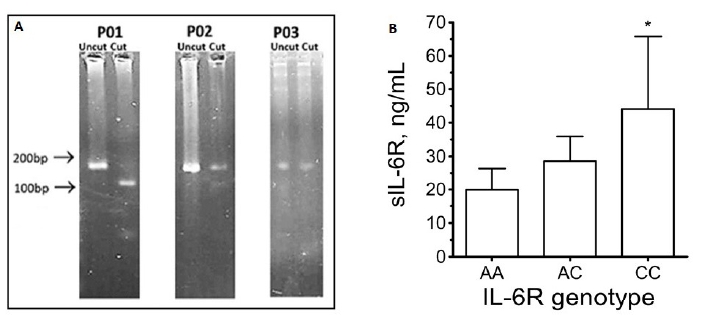
Primers flanking the rs2228145 site were designed using Primer-BLAST software (https://www.ncbi.nlm.nih.gov/primer-blast): Forward: 5’-TGACAGCACCAGCTAAGT-3’; Reverse: 5’-ACAATGGCAATGCAGAGGAG-3’. Primers plus 1.78µg DNA template were included within 25µl PCR reactions (95oC/3min; 35 cycles of 95oC/30s, 61oC/30s, 72oC/60s; 72oC/5min). PCR products were subjected to HinfI restriction digests (5µl CutSmart buffer (10x); 1µl HinfI; 1µg DNA, within 50µl reaction volumes; 37oC; 60min). Given the presence of the HinfI consensus sequence (GATTC) in PCR product from AA homozygoytes, but its absence in CC homozygotes (GCTTC), samples’ banding patterns in agarose-gels (Fig1A) reflected the genotype of the sample donor: either an intact PCR product (183bp [CC homozygotes]), a pair of fragments (101bp and 82bp [AA homozygotes]), or a triplet of bands (183bp, 101bp, and 82bp [AC heterozygotes]).
International Physical Activity Questionnaires (IPAQ)
Prior to sample donation, all participants completed IPAQs to estimate self-selected PA levels in the week preceding sample donation. In each case, IPAQ results were processed to produce a total PA score (MET-mins/week).
Statistical Analysis
All statistical analyses were performed using MiniTab19 software (MiniTab Ltd, Coventry, UK). Shapiro-Wilks tests were carried out to determine whether parameters were normally-distributed. 2-sample t-tests or 1-way ANOVA (plus Dunnett’s post-hoc comparisons) were used for analysis of normally-distributed parameters within two-sample or multiple-sample datasets, respectively. Because IPAQ was not normally distributed, non-parametric analyses (Mann-Whitney or Mood’s median test) were carried out for two-sample or multiple-sample datasets, respectively. Regression analysis was used to evaluate potential associations between two numerical parameters within the same dataset. Data were expressed as mean±standard deviation, with significance set at P<0.05.
RESULTS
Group-1: Agarose-gel banding patterns revealed that the cohort comprised 6 AA homozygotes (50.0%), 4 AC heterozygotes (33.3%), and 2 CC homozygotes (16.7%), which showed significant similarity with previously-reported rs2228145 genotype frequencies (eg. International HapMap Project [2007; https://www.genome.gov/10001688/international-hapmap-project]). Also in agreement with previous reports [3-6], sIL-6R levels were in the 20-80ng/ml range, with significant genotype-dependent differences in blood-borne sIL-6R levels (CC (44.1±21.7ng/mL) vs. AC (28.6±7.3ng/mL) vs. AA (19.9±6.5ng/mL); Fig 1B).
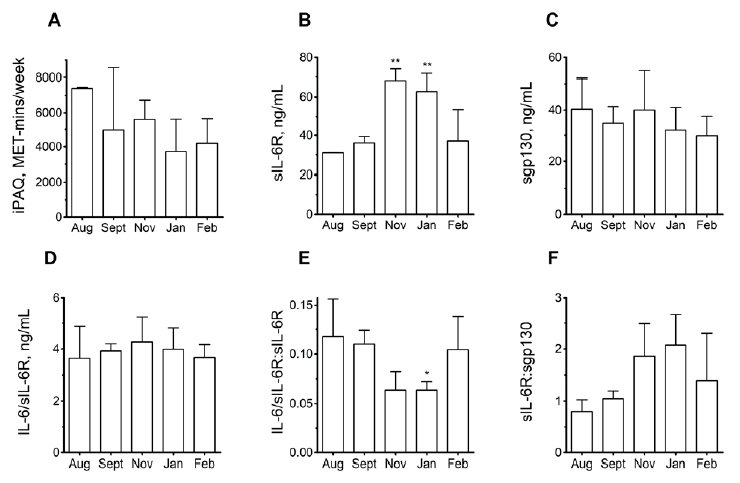
Group-2: PA levels of the participants (2 AA homozygotes; 4 AC heterozygotes) did not significantly vary during the 6-month sampling period, although non-significant trends towards lower PA in January compared to August were apparent (Fig2A). Conversely, sIL-6R levels were significantly higher in the November-January period than in August (Fig2B). As for Group-1, AA homozygotes had lower plasma sIL-6R levels than AC heterozygotes across the entire 6-month period, albeit without this difference achieving statistical significance [data not shown]. No monthly differences in sgp130 or IL-6/sIL-6R were observed (Figs2C-D), but the proportion of sIL-6R complexed with IL-6 (ie. in ‘active complexes’) underwent a significant decrease in January (Fig2E). The proportion of sIL-6R complexed with sgp130 (ie. in ‘inactive complexes’) underwent a non-significant increase (P=0.096) in January (Fig2F). Importantly, when average PA readings from across the 6-month time-window were calculated, AA homozygotes undertook significantly more PA than AC heterozygotes (6318±899 v. 3904±2280 MET-mins/week; Fig3A).
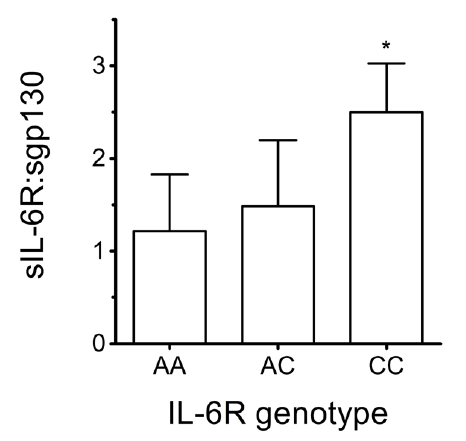
Overall: Data from the two cohorts were pooled into a composite dataset (age=30±12yrs; 12 AA homozygote samples [40.0%]; 16 AC heterozygote samples [53.3%]; 2 CC homozygote samples [6.7%]). Again, these observed frequencies showed significant similarity to previously-reported rs2228145 frequencies. Ratios of IL-6/sIL-6R complex-to-total sIL-6R were low across all three genotypes (4.1±1.1ng/ml [complex] vs. 39.4±18.9ng/ml [sIL-6R]), indicating that only ~10% of sIL-6R molecules were in ‘active complexes’. This was expected, as IL-6 release as a myokine should be minimal given that participants had refrained from exercising for >12hrs before sampling points [2]. In contrast, sIL-6R:sgp130 ratios were significantly higher for CC than for AA homozygotes (Fig4).
Weekly PA levels showed similar genotype-dependent variation as those seen for Group-2 (P<0.05; data not shown). Non-significant inverse correlations were observed between plasma sIL-6R and PA levels (P=0.197, r -0.25; Fig3B), and between plasma sgp130 and PA levels (P=0.160, r=-0.27; data not shown). We and others [4,11] have previously observed similar inverse correlations, which have been tentatively attributed to exercise-associated regulatory mechanisms controlling either expression of proteases responsible for sIL-6R ‘shedding’ [12], or influx of leukocyte subpopulations (as sources of sIL-6R ‘shedding’) into the vicinity of damaged muscle tissue post-exercise [4].
DISCUSSION
Genomic markers linked to biological processes relevant to exercise-associated health benefits and/or sporting performance are used to guide decision-making in healthcare and sport/exercise science [13]. This project aimed to investigate whether the IL-6R gene (and specifically one’s rs2228145 genotype) can be provisionally designated a novel marker for PA/exercise tolerance.
Longitudinal data showed low PA levels coinciding with high sIL-6R levels in November and January, while the converse was the case during periods when PA was higher. This appears to support our previous proposal that sIL-6R could represent a marker of athletes’ exercise/training levels that is sensitive to changes on weekly/monthly bases [11], and to extend this principle by applying it to cohorts of non-elite athletes.
In contrast to previous studies (which reported that sgp130 levels exceed those of sIL-6R [3]), our data indicates that sIL-6R and sgp130 are present in capillary blood at approximately equivalent levels (see Fig2). Importantly, the genotype-dependent pattern seen in Fig4 suggests sIL-6R:sgp130 ratios of ~1:1 for AA, but ~2:1 for CC. Given the hexameric 2:2:2 IL-6:sIL-6R:sgp130 stoichiometry of the ‘inactive complex’ [3], our data suggest that lower proportions of sIL-6R molecules may be bound within ‘inactive complexes’ in CC individuals (thus implying greater potential for trans-signalling). Additionally, decreased mbIL-6R cell-surface expression, and hence decreased responsiveness to classical signalling, have been associated with the CC genotype [14]. This suggests that the balance of trans- versus classical-signalling may differ in a genotype-dependent manner (ie. favouring trans-signalling in C-allele bearers). Importantly, this may provide a mechanism underpinning the genotype-dependent variation in PA levels illustrated in Fig3A.
As stated earlier, associations between blood-borne sIL-6R levels and ‘over-reaching’ [11] suggest that rs2228145 genotype may influence participation in exercise, perhaps by impacting on one’s ability to tolerate PA without experiencing adverse effects. As an exploratory pilot-study, the current study has numerous limitations (eg. small cohort sizes; reliance on questionnaire responses concerning self-reported PA levels; no recording of participants’ perceptions regarding undertaking of exercise). Nevertheless, we hope that the current study provides a starting-point from which future investigations may confirm our initial findings; determine whether rs2228145 genotype is associated with severity of over-reaching symptoms; and investigate which symptoms are most affected, and why?
Future studies could also investigate specific physiological contexts in which rs2228145 genotype may impact on exercising participants’ wellbeing. For example, sIL-6R’s ability to cross the blood-brain barrier and elicit neuroinflammatory trans-signalling responses in CNS cells has been linked to cognitive responses that present as fatigue, reduced power output, and termination of exercise [15]. Interestingly, we have previously reported that exercising participants’ self-reported sensations of fatigue have been reported to correlate with their plasma sIL-6R levels [11]. Thus, it may be that triggering of such neuroinflammatory signalling, and possibly perceptions of fatigue, are more marked in individuals such as CC homozygotes with high blood-borne sIL-6R levels, and this may influence their choices regarding incorporation of exercise into their lifestyles. Similarly, rs2228145 significantly impairs lung function (specifically FVC and FEV) in CC homozygotes with asthma [10]. While these effects failed to reach statistical significance in non-asthma sufferers [10], one may speculate that weaker sub-clinical effects at rest may pre-dispose non-asthmatic CC homozygotes to episodic upper respiratory symptoms when exercising, and so discourage participation in exercise.
CONCLUSIONS
In conclusion, this preliminary study’s findings suggest associations between rs2228145 genotype, sIL-6R, and self-selected PA levels in healthy adults. Although more research is required to confirm and extend the above findings, designation of IL-6R as a novel marker gene, and determination of clients’ rs2228145 genotypes, may in future aid design of personalised exercise programmes matched to exercise-providers’ clients, in both elite sport and exercise-referral settings [13].